Testing for SARS-CoV-2
The FLARE Four
- Two major types of assays are used to detect respiratory viral infections: Reverse-Transcriptase Polymerase Chain Reaction (RT-PCR) and Direct Fluorescent Antibody (DFA); RT-PCR continues to be the method of choice for detecting SARS-CoV-2 due to its wider availability and high sensitivity
- True positive predictive value and negative predictive value for SARS-CoV-2 RT-PCR remain unknown this early in the COVID-19 pandemic. Without a clear gold standard test, clinical sensitivity and specificity are difficult to determine
- Sampling, anatomic location, and disease stage and severity likely play critical roles in determining the sensitivity and specificity of PCR testing
- Serological tests for COVID-19 promise to supplement PCR based strategies testing
Many people are saying...different things about the sensitivity of testing for SARS-CoV-2.
Subscribe to the latest updates from FLARE Advances in Motion
How Do We Usually Detect Viral Respiratory Infections?
There are many different ways to detect a virus in a clinical or research sample. These methods generally fall into three categories: (1) detecting genetic sequences unique to the virus, (2) detecting antigens unique to the virus and (3) detecting and growing a live virus.
1. Reverse-Transcriptase Polymerase Chain Reaction (RT-PCR)
This method uses a primer pair or several primer pairs that recognize virus-specific genetic sequences and initiate an amplification process that can only begin in the presence of viral RNA. This method is highly sensitive because amplification can begin with a small amount of viral RNA, and very specific because the viral sequences used for initiation of amplification are chosen, in part, based on their absence from the human genome and other infectious agents. In fact, this method is used clinically to detect Influenza A and B, and RSV in clinical samples, and the Biofire assay can detect as many as 20 different viral pathogens using an RT-PCR-based approach.
There are also many recently developed and emerging techniques that detect small amounts of unique sequence inside a fixed and permeabilized cell (ISH, snFISH, PLISH, etc.). These methods are labor-intensive and are likely difficult to scale for clinical application at this time.
2. Direct Fluorescent Antibody (DFA) Labelling, Also Called Direct Immunofluorescence
In this approach, an antibody is raised against an antigen on the virus and conjugated to a reporter molecule such as a fluorescent molecule. When this conjugated antibody binds its target viral antigen, the sample with the virus becomes detectable by a method specific to the reporter (e.g. fluorescence microscopy). This method is used clinically to rapidly detect Adenovirus antigen, Parainfluenza 1-3 antigen and Human Metapneumovirus. A similar immunochromatography assay is used to rapidly detect Influenza A and B.
3. Recovery and Amplification of Live Virus from a Clinical Specimen
This is the most rigorous proof of viral infection. Such a test requires a specialized cell culture platform to grow the virus, takes a long time relative to other methods, and is labor-intensive. Thus, it is difficult to scale in any clinical setting. However, this approach may be required to distinguish the presence of a live virus that carries a risk of infection from the benign presence of viral RNA.
Understanding Positive and Negative Predictive Value
The sensitivity and specificity provide information about the utility of a test at the population level—what proportion of all people with the virus will we find with the test? How many people will we label incorrectly with false positive tests? In contrast, the positive predictive value (PPV) and negative predictive value (NPV) provide utility in the direct clinical care of a patient. Given a positive (or negative) test result, what is the likelihood the patient has the virus? Both positive and negative predictive values are influenced by the prevalence of the disease in the population. Due to scarcity and imperfect nature of testing, the prevalence of SARS-CoV-2 in our population remains unknown, limiting our ability to determine the true NPV and the PPV of any SARS-CoV-2 test.
Let’s take a brief statistical detour to discuss positive predictive value (PPV), negative predictive value (NPV), sensitivity (SN), and specificity (SP) (see Frank Harrell's biostatistics site for more). All probabilities are conditional on something—knowing that “something” is key to understanding them. For example: Consider women who undergo breast cancer screening mammography. Sensitivity, or true positive rate, is asking: “Given a patient is known to have breast cancer (by a gold standard), what is the probability that their mammogram will be ‘positive’?” Specificity, or true negative rate, is asking: “Given a patient is known to not have breast cancer, what is the probability that the mammogram will be ‘negative’?” These describe the technical abilities of the test to detect a specific abnormality. But we don’t do a test because we already know our patient has a disease. Clinicians care about the exact opposite question! We need to know the probability of the unknown, given the known—in clinical scenarios this is the probability of disease given the test result in front of us.
A patient with a positive mammogram needs to know: “Given the test is positive, what is the probability that she has breast cancer.” This is the positive predictive value. Similarly: Given a test is negative, what is the probability that the patient does not have the disease—negative predictive value. This takes into account the prevalence of the disease and thus pre-test (prior) probability.
What are the Options for Detecting a SARS-CoV-2 Infection?
Early in the COVID-19 outbreak, the sequence of the virus became publicly available, and RT-PCR testing for SARS-CoV-2 became the standard approach for testing (see figure below). To measure the amount of viral RNA in the sample, two nucleic acid primers are designed to bind specifically to genetic sequences in the SARS-CoV-2 virus (after reverse transcription) and nowhere else in the human genome or in other known viral genomes. The subsequent PCR cycle-based DNA amplification to a pre-specified threshold of DNA molecules is determined entirely by the number of starting viral template copies. Thus, the real time RT-PCR cycle threshold (Ct) becomes the surrogate for viral load in the sample. A lower number of cycles indicates a higher viral load, since fewer cycles were needed to amplify RNA to pre-validated number of PCR products.
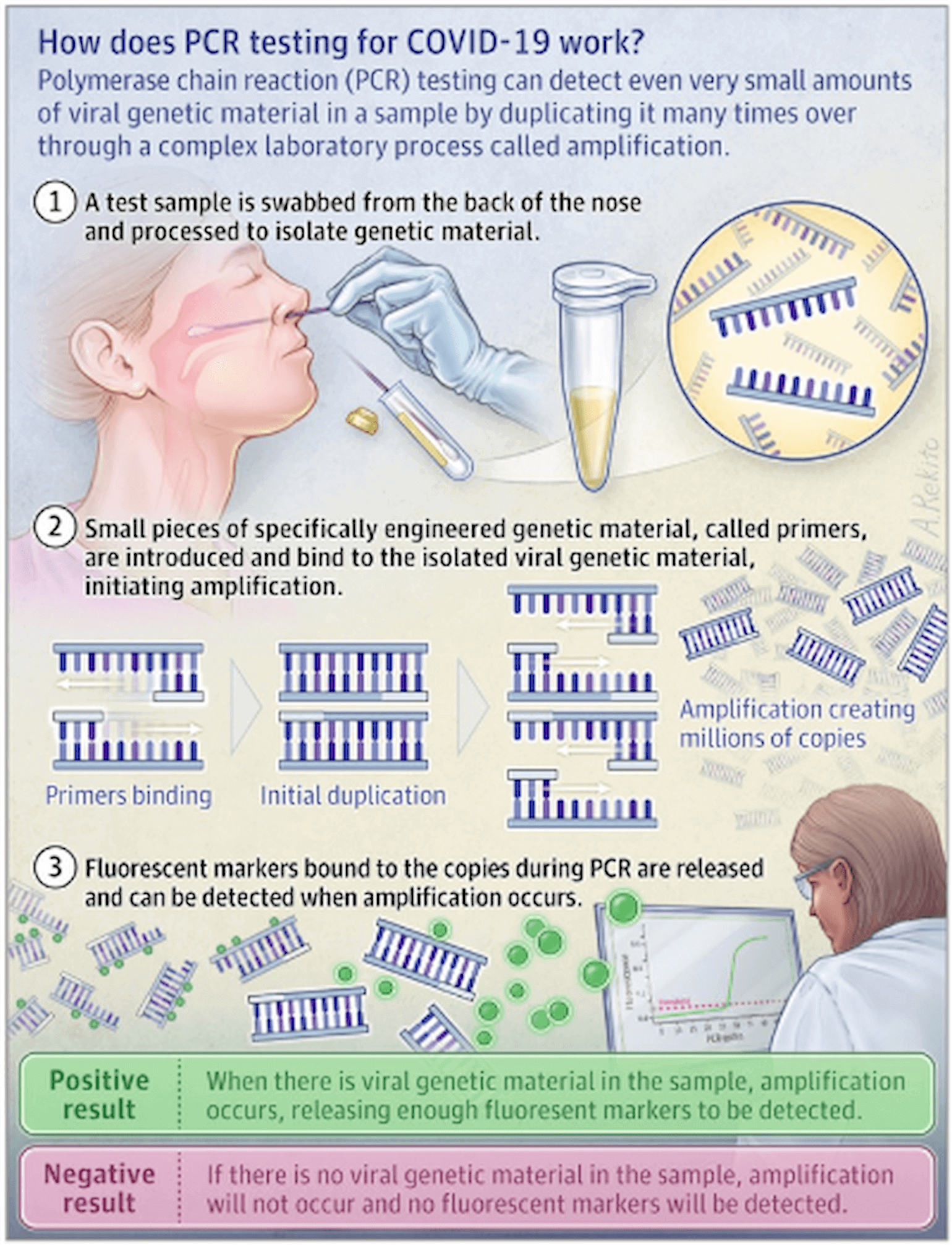
Figure 1
(Hadaya et al. JAMA 2020)
DFA-based tests are currently under development. These tests are typically much faster than RT-PCR. However, while they may be highly specific, at this time, their sensitivity is not sufficiently high to make them clinically useful. During an outbreak, it is vital that patients who test negative are truly negative. Thus the negative predictive value of a test is much more important than its positive predictive value.
Serologic Testing for SARS-CoV-2 Infection
An alternative approach to testing involves not detection of the virus or its antigens but antibodies directed against those antigens. Antibody tests can be used to determine the efficacy of vaccines, identify donors for convalescent plasma (March 27 FLARE), confirm past infections, and reveal epidemiologically how many people have been exposed. Serological testing may also prove to be useful in clinical cases where direct viral testing is indeterminate.
In prior studies on SARS-CoV, development of IgG against viral antigens was seen at a median 17 days, and IgM was seen at 20 days (Chan et al., 2005; Woo et al., 2004). This is shown below.
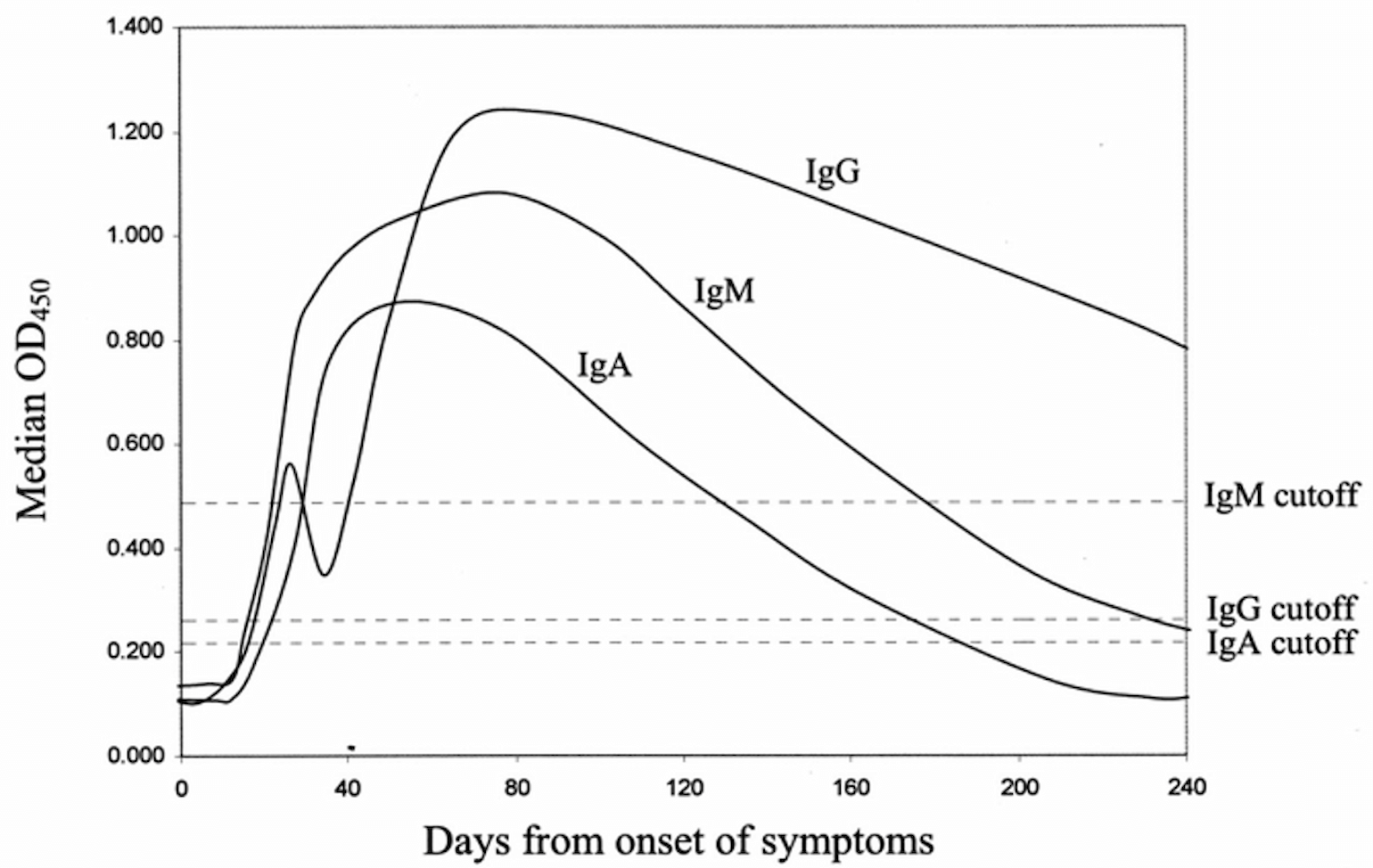
Figure 2
Days from onset of symptoms: Development of IgG against viral antigens was seen at a median 17 days, and IgM was seen at 20 days (Chan et al., 2005; Woo et al., 2004).
As SARS-CoV-2 is similar to SARS-CoV, it is believed that it will incite a similar immunologic response. To evaluate the immune response in SARS-CoV-2, Zhao et al. conducted a study which enrolled 173 patients with confirmed COVID-19. IgM was seen at a median of 12 days and IgG at 14 days. In total, 171 patients (93.1%) had antibodies of some type detected; 2 of the 173 patients were tested early in their course and did not have antibody detected. The sensitivity in days 8-15 from symptom onset was 89.6% for total antibody (antibodies of any type, including IgG and IgM), 73% for IgM and 54.1% for IgG. When combined with a RT-PCR test, sensitivities improved to 97-100% after day 8 (Zhao et al., 2020).
A point of care lateral flow immunoassay test was developed in China to test both IgM and IgG in blood. Using RT-PCR as the gold standard, they found a sensitivity of 88.66% and a specificity of 90.63% (Li et al., 2020). False positive results may be due to the imperfect sensitivity of the PCR-based tests and could have been true positive, or due to cross reactivity of antibody test to other coronaviruses. Overall, immunoassays show promise to be used in combination with RT-PCR to improve testing reliability.
What is the Best Sample to Detect SARS-CoV-2 for Real Time RT-PCR?
A major factor in determining test sensitivity is the anatomic location of sample collection. In a non-peer-reviewed paper, 213 patients identified to have COVID-19 in China had 866 samples collected from multiple respiratory sources. Patients were additionally classified by their duration of symptoms and severity of disease. Overall, BAL fluid yielded the highest % positive, but this was only obtained in patients with severe disease later in the course of infection. For patients early in the course of the disease, sputum sample performed better than nasal and throat swabs.
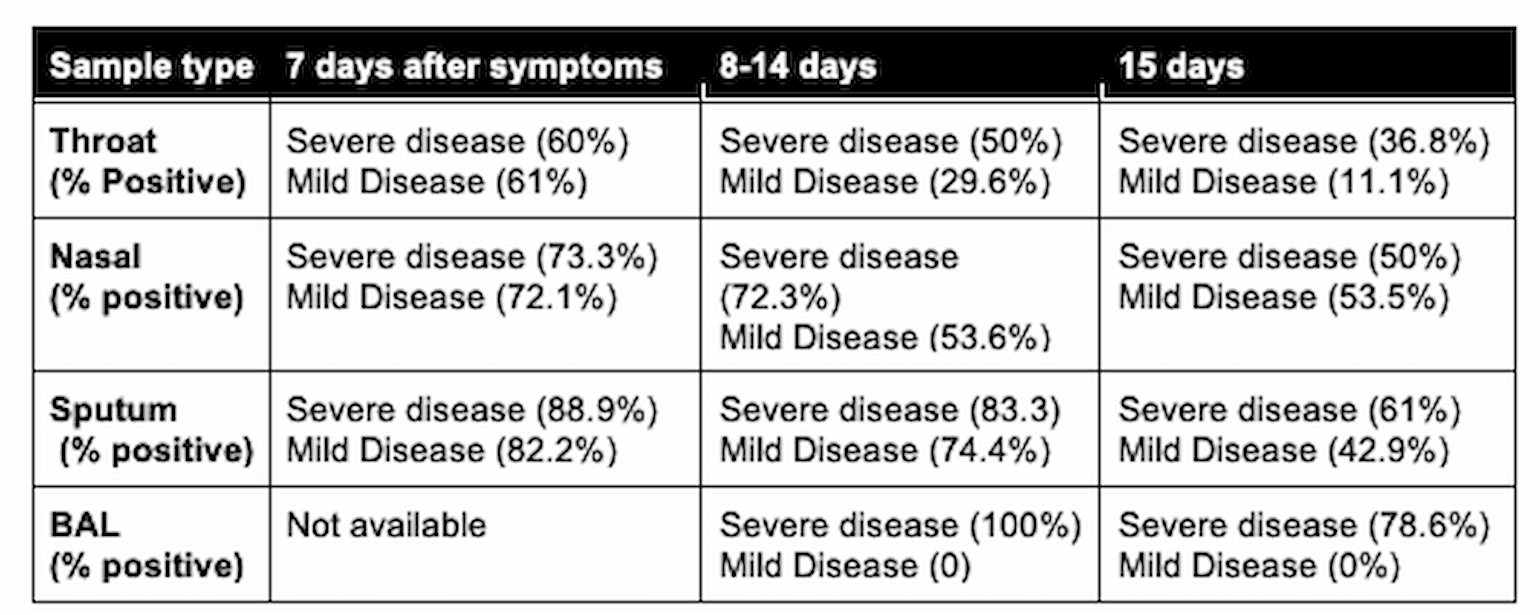
Table 1
* severe disease defined as dyspnea, respiratory frequency ≥30/minute, blood oxygen saturation ≤93%, PaO2/FiO2 ratio <300, and/or lung infiltrates >50% of the lung field within 24-48 hours by guidelines of 2019-nCoV infection of the National Health Commission of the People’s Republic of China (Yang et al., 2020).
In another study, 1070 specimens from 205 patients with confirmed COVID-19 were similarly evaluated with samples taken from different body sites. The results are summarized here:
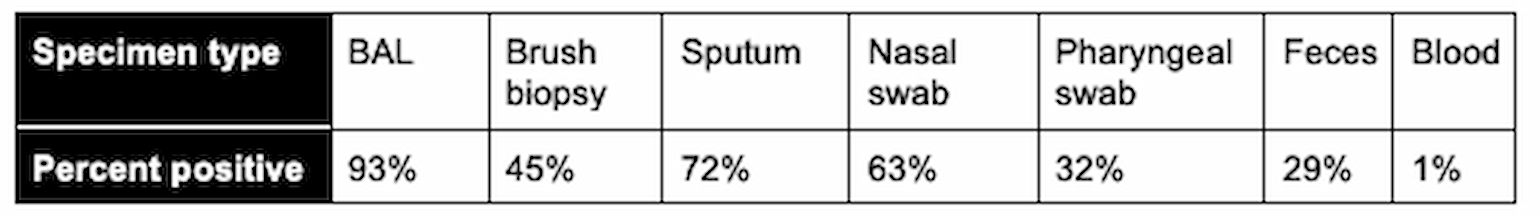
Table 2
(Wang et al., 2020)
Finally, Zou et. al. directly compared the viral load in nasal and oropharyngeal swabs from the same 18 patients at different times during the disease by RT-PCR (Zou et al., 2020). In general, higher viral loads were detected earlier in the disease course, and in samples collected from the nasopharynx had higher sensitivity compared to the oropharynx. Other studies similarly evaluated the viral load over time and space. One study showed that viral load in early stages was significantly higher than that in recovery stages (Yu et al., 2020). Salivary viral load was highest during the first week since symptoms started. Older age was correlated with higher viral load (To et al., 2020).
Thus, the specimen type, time of testing in the course of the patient’s illness and perhaps even patient-specific factors influence the likelihood of recovering viral RNA, which, in turn, determines the positive and negative predictive value of these tests. Nasopharyngeal (NP) swab balances relatively high sensitivity with relatively easy sample access both early and late in the disease. Currently it is important to note that NP swabs are run “in house” at the MGH lab. Sputum and tracheal aspirates, which may have higher sensitivity, are currently sent to the MA state lab, resulting in much longer turnaround time.
In Summary
Clinical sensitivity and specificity of a viral test depends on the testing procedure itself (PCR vs. serology) but also on specimen type, anatomic location of the sample, depth of the swab (for some anatomic locations), and probably factors in sample quality control and processing (including time from collection to RNA extraction). Test performance may also depend on the stage, severity, and symptoms of the viral disease. All of these factors lead to a situation in which no single test can give a 100% reliable answer. Based on the above data, NP swab and PCR provide the best sensitivity and specificity, but positive and negative predictive value will, again, depend on the prevalence in the population being tested (or pre-test probability of the patient). The unknown prevalence, along with sample collection issues documented above, continue to limit our ability to answer the clinically vital question of: does this patient have SARS-CoV-2? Because this question has public health as well as clinical implications, it will likely have to be addressed via a combined approach - i.e. PCR + antibody testing.
References
- Chan, K.H., Cheng, V.C.C., Woo, P.C.Y., Lau, S.K.P., Poon, L.L.M., Guan, Y., Seto, W.H., Yuen, K.Y., and Peiris, J.S.M. (2005). Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross-reactivity with human coronaviruses 229E, OC43, and NL63. Clin. Diagn. Lab. Immunol. 12, 1317–1321.
- Li, Z., Yi, Y., Luo, X., Xiong, N., Liu, Y., Li, S., Sun, R., Wang, Y., Hu, B., Chen, W., et al. (2020). Development and Clinical Application of A Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med. Virol.
- To, K.K.-W., Tsang, O.T.-Y., Leung, W.-S., Tam, A.R., Wu, T.-C., Lung, D.C., Yip, C.C.-Y., Cai, J.-P., Chan, J.M.-C., Chik, T.S.-H., et al. (2020). Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis.
- Wang, W., Xu, Y., Gao, R., Lu, R., Han, K., Wu, G., and Tan, W. (2020). Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA.
- Woo, P.C.Y., Lau, S.K.P., Wong, B.H.L., Chan, K.-H., Chu, C.-M., Tsoi, H.-W., Huang, Y., Peiris, J.S.M., and Yuen, K.-Y. (2004). Longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin. Diagn. Lab. Immunol. 11, 665–668.
- Yang, Y., Yang, M., Shen, C., Wang, F., Yuan, J., Li, J., Zhang, M., Wang, Z., Xing, L., Wei, J., et al. (2020). Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. Preprints.
- Yu, F., Yan, L., Wang, N., Yang, S., Wang, L., Tang, Y., Gao, G., Wang, S., Ma, C., Xie, R., et al. (2020). Quantitative Detection and Viral Load Analysis of SARS-CoV-2 in Infected Patients. Clin. Infect. Dis.
- Zhao, J., Jr., Yuan, Q., Wang, H., Liu, W., Liao, X., Su, Y., Wang, X., Yuan, J., Li, T., Li, J., et al. (2020). Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019 (medRxiv).
- Zou, L., Ruan, F., Huang, M., Liang, L., Huang, H., Hong, Z., Yu, J., Kang, M., Song, Y., Xia, J., et al. (2020). SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 382, 1177–1179.
View all COVID-19 updates
Learn about research in the Division of Pulmonary and Critical Care Medicine