Cats and Bats and Pangolins: The Origin of Sars-Cov-2
The FLARE Four
- SARS-CoV-2 shares 96% whole genome identity with a coronavirus found in bats, BatCoV RaTG13
- The key differences between SARS-CoV-2 and BatCoV localize to the receptor binding domain (RBD) of the spike protein, required for engaging the host receptor ACE2
- The RBD of SARS-CoV-2 is closely related to a coronavirus found in pangolins near the origin of the pandemic in China. It is therefore extremely likely that SARS-CoV-2 emerged from a recombination event between bat and pangolin virus, though the exact nature of the event is hard to determine
- There is nothing to suggest SARS-CoV-2 is human-made
Many people are asking...where did SARS-CoV-2 come from? “You have heard all different things. Three or four different concepts as to how it came out.”
Subscribe to the latest updates from FLARE Advances in Motion
Introduction
Consensus scientific opinion is that SARS-CoV-2 emerged via a recombination event between related animal coronaviruses. The strong evidence for this conclusion has not precluded a great deal of speculation that the virus either was engineered or somehow ”escaped“ from a laboratory. In tonight’s FLARE, we review what is known about the SARS CoV-2 genome and what it tells us about the virus’ likely origin story.
The SARS-CoV-2 Genome
In order to understand the evidence for a natural origin of SARS-CoV-2, it is first necessary to review a bit of viral biology. SAR-CoV-2 is a single strand “plus” (or “positive-sense”) RNA virus of the coronavirus family. Coronaviruses (CoVs) comprise a diverse family which infect a variety of animals including, of course, human beings. CoVs are subdivided into 4 genera - alpha, beta, gamma, and delta. Alphacoronaviruses and betacoronaviruses infect mammals, while the gamma and delta genera typically affect birds. These viruses can be organized into a phylogenetic tree (analogous to a family tree) based upon genome sequence similarity. SARS-CoV-2, like MERS-CoV and SARS-CoV, is a betacoronavirus. The betacoronavirus genus is further subdivided into 5 subgenera. SARS-CoV-2 and SARS-CoV are of the Sarbecovirus subgenus, but of different clades (branches of phylogenetic tree with a common ancestor).
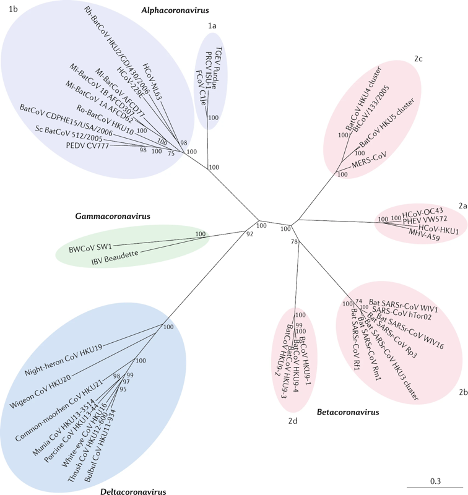
Figure 1
Coronavirus diversity as divided into a phylogenetic tree (Cui, Li, and Shi 2019).
Viral Infection and Cell Entry
Although betacoronavirus clades are defined by differences in the key gene RNA-dependent RNA polymerase (RdRp), important information about viral origin may also be obtained by examining sequence variation in the spike (”S”) protein - a critical actor in viral entry and a target of vaccine efforts (covered by FLARE on April 24th). While still the topic of some debate, there appear to be at least two independent mechanisms by which SARS-CoV-2 enters host cells (see March 28 FLARE):
- Via fusion: The receptor binding domain (RBD) of the viral S-protein attaches to ACE2 protein on the cell surface of the host cell. The RBD is made up of a core structure and a receptor binding motif (RBM) which governs the physical interaction with ACE2. The RBD of SARS-CoV and SARS-CoV-2 share ~75% sequence homology, while the RBMs share ~50% homology (Wan et al. 2020). Cell entry further requires that the S-protein be cleaved by a host-cell protease at the S1/S2 and S2’ cleavage sites to facilitate membrane fusion and viral entry. This process is well-described in SARS-CoV-1. Of note, SARS-CoV-2, unlike SARS-CoV-1, has a furin protease recognition motif in the S-protein. The furin protease is widely expressed by host cells - potentially increasing viral infectivity (Shang et al. 2020) A furin cleavage site is found in many human proteins, and renders the S1/S2 site susceptible to cleavage by any one of a large family of proteases (PCSK1-PCSK9), including Furin (PCSK3) (Braun and Sauter 2019). This expansion of the host protease repertoire may in part explain the apparent higher efficiency of infection of SARS-CoV-2 compared to SARS-CoV-1 (Hoffmann, Kleine-Weber, and Pöhlmann 2020).
- Via endocytosis: Under conditions where S-protein mediated viral envelope fusion does not occur, coronaviruses may also be internalized by way of receptor mediated endocytosis. Through this process the intact virus is encapsulated within an endosome, from which it then escapes by way of sequential endosome acidification, lysosomal fusion, and cathepsin-mediated S-protein processing.
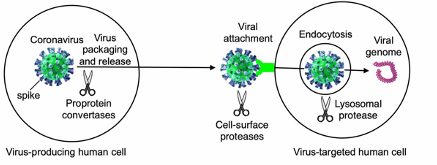
Figure 2
Different stages of coronavirus entry where host cellular proteases may activate coronavirus spikes (Shang et al. 2020).
So Where Did SARS-CoV-2 Come From? And What is This Business About Pangolins?
Many pathogenic coronaviruses reach their eventual victims by way of a definitive host and an intermediate host. In the case of SARS and MERS, sequence analysis revealed bats to be definitive hosts in both cases. Civet cats served as intermediate hosts for SARS-CoV while camels did so for MERS-CoV. This history made it likely that a similar pattern of hosts would exist for SARS-CoV-2.
Clues as to the origins of SARS-CoV-2 come primarily from analyses of genetic homology to known animal coronaviruses - particularly from study of the S protein receptor binding domain (RBD) and S1/S2 cleavage site. SARS-CoV-2 shares 96% genome identity with a coronavirus found in bats, BatCoV RaTG13 - highly suggestive of an origin for the human pathogen in bats. There are, however, key differences between SARS-CoV-2 and RaTG13 both in the RBD and at the S1/S2 cleavage site. What might account for these?
The subgenus Sarbecovirus (which counts among its members SARS-CoV and SARS-CoV-2) is well-appreciated to undergo recombination events. Notably, the RBD of SARS-CoV-2 is extremely close in sequence to that of a coronavirus recently isolated from Malayan pangolins near the origin of the pandemic in China (Zhang, Wu, and Zhang 2020). Specifically, metagenomic sequencing has identified a sublineage of pangolin-associated coronavirus (Lam et al. 2020) harboring a single amino acid difference as compared to SARS-CoV-2 RBD. Moreover, the 5 critical RBD residues for ACE2 binding are identical between the two (Xiao et al. 2020; Zhang, Wu, and Zhang 2020). The likelihood of the pangolin CoV and SARS-CoV-2 separately developing such similar structures entirely by chance is incredibly low and suggests instead that this occurred either via parallel natural selection or (more likely) by recombination of the virus between the bat and ancestral pangolin isolates (Andersen et al. 2020) (Figure 4).
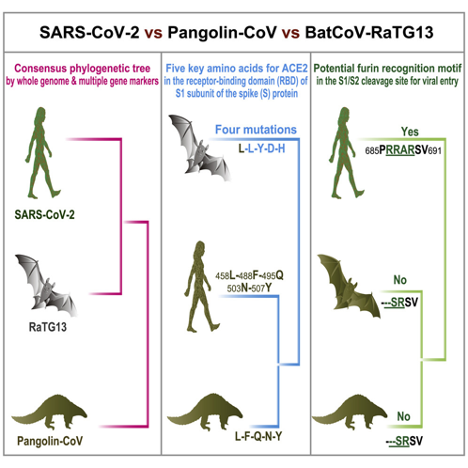
Figure 4
Potential evolution of a SARS-CoV-2 virus (Zhang et al. 2020).
Moreover, studies have also uncovered evidence of apparent pathogenic infection in pangolins, noting “diffuse alveolar damage of various severity in the lung … alveoli were filled with desquamated epithelial cells and some macrophages with hemosiderin pigments, with significantly reduced alveolar space, leading to the consolidation of the lung” (Xiao et al. 2020). It is therefore believed that one or more bat coronaviruses infected pangolins and thereafter underwent multiple recombination events with other CoVs (Hon et al. 2008; Lam et al. 2020) to produce the ancestors of SARS-CoV-2. Importantly, and unlike for SARS-CoV isolated from civets, substantial sequence differences (outside the RBD) between SARS-CoV-2 and Pangolin-CoV imply that the pangolin pathogens isolated to date are not directly responsible for human infection. Whether these observed differences reflect yet unidentified additional pangolin strains awaits further study.

Figure 5
A pangolin, prior to developing Coronavirus-related ARDS (from https://www.worldwildlife.org/species/pangolin).
So Was SARS-CoV-2 Made in a Lab?
The homology between bat coronavirus, pangolin coronavirus and SARS-CoV-2 provides compelling evidence of natural origin. Left unexplained is the presence of the furin cleavage site which appears in neither antecedent virus. There are 2 main scientific theories to explain how the extant SARS-CoV virus came into existence and one comprehensively debunked theory: 1) natural selection in animal hosts prior human infection and 2) natural selection in humans after the virus transferred to humans. The debunked theory suggested that the virus came from a lab (Andersen et al. 2020).
Emergence of SARS-CoV RBD and Furin Cleavage Sites by Natural Selection
None of the pangolin CoV isolates identified to date harbor the S1/S2 furin-mediated multibasic cleavage site in the S-protein. In fact, all related coronaviruses have a monobasic cleavage site (Lam et al. 2020). How the furin-mediated multibasic cleavage site was incorporated in the viral genome is not currently understood. The region of the S1/S2 junction is a hotspot for mutation (Yamada and Liu 2009) and thus the virus could certainly have acquired and maintained such a mutation in, for example, pangolins if it were adaptive.
It is also possible that an ancestor of SARS-CoV-2 circulated for some time in humans before undergoing natural selection to become the virus we know today. As the bat-CoV RBD renders it unable to infect humans, it is likely such a progenitor would have come from a pangolin as outlined above (Lam et al. 2020; Xiao et al. 2020). In this model, the insertion of the polybasic furin-dependant cleavage site occurred later, during the human-to-human transmission stage of the virus. This would imply that there were likely human cases prior to November 2019 (Bryner 2020) which went unrecognized and which may have been mild clinically. If the virus passed from animals to humans multiple times (as for MERS-CoV) this model predicts the yet unrecognized possible existence of multiple short transmission chains prior to the evolution of the cleavage site (Andersen et al. 2020).
Generation of SARS-CoV RBD and Furin Cleavage Sites by Artificial Selection
Could the SARS-CoV-2 RBD and furin cleavage site have arisen during cell culture or in animal models in a laboratory? Coronavirus research has been happening for many years and there have been past instances in which SARS has accidentally been released (Lim et al. 2004). It is formally possible that the RBD mutated from a SARS-CoV-like bat virus in the lab to the observed SARS-CoV-2 RBD, but this theory fails to account for the fact that a nearly identical RBD is found naturally in the pangolin. It is therefore much much more likely the RBD changes arose from recombination or mutation via natural selection. A ”lab evolution” explanation also would not easily explain the co-presence of the furin-mediated multibasic cleavage site - a feature only ever reported in culture with passage of avian flu (Ito et al. 2001).
Other lines of evidence argue against a laboratory origin as well. If the virus had been artificially mutated or engineered, we would expect to see sequence signatures of laboratory strain backbones - which have not been described. In addition, modeling studies suggest that the SARS-CoV-2 RBD is not optimally ”designed” for binding the human ACE2. Had the virus been made in a lab for the express purpose of infecting human cells, scientists would therefore likely have chosen other, higher affinity ACE2-binding motifs than present in SARS-CoV-2 (Damas et al. 2020).
Conclusions
SARS-CoV-2 is similar to bat coronavirus RaTG13, with ~96% sequence homology. Based upon similarities in the spike protein and RBD as well as epidemiological circumstantial evidence, the Malayan pangolin likely represents the critical intermediate host for SARS-CoV-2 prior to widespread human infection. It is yet unclear whether evolution of the RBD and S-protein occurred either through recombination or convergent evolution, but all evidence points to a natural rather than man-made origin. The rumor that SARS-CoV-2 was created in the lab is just a rumor.
References:
- Andersen, Kristian G., Andrew Rambaut, W. Ian Lipkin, Edward C. Holmes, and Robert F. Garry. 2020. “The Proximal Origin of SARS-CoV-2.” Nature Medicine 26 (4): 450–52.
- Braun, Elisabeth, and Daniel Sauter. 2019. “Furin-Mediated Protein Processing in Infectious Diseases and Cancer.” Clinical & Translational Immunology 8 (8): e1073.
- Bryner, Jeanna. 2020. “1st Known Case of Coronavirus Traced back to November in China.” Live Science,[available at D Https://www. Livescience. Com/first-Case-Coronavirus-Found. Html Accessed on March 31 2020]. https://www.livescience.com/first-case-coronavirus-found.html.
- Cui, Jie, Fang Li, and Zheng-Li Shi. 2019. “Origin and Evolution of Pathogenic Coronaviruses.” Nature Reviews. Microbiology 17 (3): 181–92.
- Damas, J., G. M. Hughes, K. C. Keough, and C. A. Painter. 2020. “Broad Host Range of SARS-CoV-2 Predicted by Comparative and Structural Analysis of ACE2 in Vertebrates.” bioRxiv. https://www.biorxiv.org/content/10.1101/2020.04.16.045302v1.abstract.
- Hoffmann, Markus, Hannah Kleine-Weber, and Stefan Pöhlmann. 2020. “A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells.” Molecular Cell, April. https://doi.org/10.1016/j.molcel.2020.04.022.
- Hon, Chung-Chau, Tsan-Yuk Lam, Zheng-Li Shi, Alexei J. Drummond, Chi-Wai Yip, Fanya Zeng, Pui-Yi Lam, and Frederick Chi-Ching Leung. 2008. “Evidence of the Recombinant Origin of a Bat Severe Acute Respiratory Syndrome (SARS)-like Coronavirus and Its Implications on the Direct Ancestor of SARS Coronavirus.” Journal of Virology 82 (4): 1819–26.
- Ito, T., H. Goto, E. Yamamoto, H. Tanaka, M. Takeuchi, M. Kuwayama, Y. Kawaoka, and K. Otsuki. 2001. “Generation of a Highly Pathogenic Avian Influenza A Virus from an Avirulent Field Isolate by Passaging in Chickens.” Journal of Virology 75 (9): 4439–43.
- Lam, Tommy Tsan-Yuk, Marcus Ho-Hin Shum, Hua-Chen Zhu, Yi-Gang Tong, Xue-Bing Ni, Yun-Shi Liao, Wei Wei, et al. 2020a. “Identifying SARS-CoV-2 Related Coronaviruses in Malayan Pangolins.” Nature, March. https://doi.org/10.1038/s41586-020-2169-0.
- Lim, Poh Lian, Asok Kurup, Gowri Gopalakrishna, Kwai Peng Chan, Christopher W. Wong, Lee Ching Ng, Su Yun Se-Thoe, et al. 2004. “Laboratory-Acquired Severe Acute Respiratory Syndrome.” The New England Journal of Medicine 350 (17): 1740–45.
- Shang, Jian, Yushun Wan, Chuming Luo, Gang Ye, Qibin Geng, Ashley Auerbach, and Fang Li. 2020. “Cell Entry Mechanisms of SARS-CoV-2.” Proceedings of the National Academy of Sciences of the United States of America, May. https://doi.org/10.1073/pnas.2003138117.
- Wan, Yushun, Jian Shang, Rachel Graham, Ralph S. Baric, and Fang Li. 2020. “Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus.” Journal of Virology 94 (7). https://doi.org/10.1128/JVI.00127-20.
- Xiao, Kangpeng, Junqiong Zhai, Yaoyu Feng, Niu Zhou, Xu Zhang, Jie-Jian Zou, Na Li, Yaqiong Guo, Xiaobing Li, Xuejuan Shen, Zhipeng Zhang, Fanfan Shu, Wanyi Huang, Yu Li, Ziding Zhang, Rui-Ai Chen, Ya-Jiang Wu, Shi-Ming Peng, Mian Huang, Wei-Jun Xie, Qin-Hui Cai, Fang-Hui Hou, Yahong Liu, et al. 2020. “Isolation and Characterization of 2019-nCoV-like Coronavirus from Malayan Pangolins.” bioRxiv. https://doi.org/10.1101/2020.02.17.951335.
- Xiao, Kangpeng, Junqiong Zhai, Yaoyu Feng, Niu Zhou, Xu Zhang, Jie-Jian Zou, Na Li, Yaqiong Guo, Xiaobing Li, Xuejuan Shen, Zhipeng Zhang, Fanfan Shu, Wanyi Huang, Yu Li, Ziding Zhang, Rui-Ai Chen, Ya-Jiang Wu, Shi-Ming Peng, Mian Huang, Wei-Jun Xie, Qin-Hui Cai, Fang-Hui Hou, Wu Chen, et al. 2020. “Isolation of SARS-CoV-2-Related Coronavirus from Malayan Pangolins.” Nature, May. https://doi.org/10.1038/s41586-020-2313-x.
- Yamada, Yoshiyuki, and Ding Xiang Liu. 2009. “Proteolytic Activation of the Spike Protein at a Novel RRRR/S Motif Is Implicated in Furin-Dependent Entry, Syncytium Formation, and Infectivity of Coronavirus Infectious Bronchitis Virus in Cultured Cells.” Journal of Virology. https://doi.org/10.1128/jvi.00613-09.
- Zhang, Tao, Qunfu Wu, and Zhigang Zhang. 2020. “Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak.” Current Biology: CB 30 (7): 1346–51.e2.
Learn more about research in the Division of Pulmonary and Critical Care Medicine
View all COVID-19 updates