Using Antibody Tests for COVID-19
The FLARE Four
- Accurate SARS-CoV-2 serology will be critical for managing the COVID-19 pandemic, allowing diagnosis of previous infection, assessment of seroprevalence, and potentially the identification of individuals with protective immunity
- Serological assays vary with regard to their sensitivity, specificity, and ability to generate quantitative versus qualitative readouts
- Technical limitations of serological assays as well as kinetics of the antibody response and pre-test probability of seropositivity must be considered when interpreting these test results
- Attempts have been made to utilize antibody testing to determine the prevalence of SARS-CoV-2 antibodies in the general population, but these studies suffer from significant methodological issues
Many people are saying...serologic testing may guide our pandemic response.
Subscribe to the latest updates from FLARE Advances in Motion
Introduction
Antibody testing holds promise as a method by which to identify the true past incidence or current prevalence of SARS-CoV-2 infection (Sood et al. 2020; NIAID n.d.). Antibody testing may also be helpful for determining infection status in a patient with a high clinical suspicion for disease but with negative viral testing, for identifying potential donors for convalescent plasma, and for assessing immunologic response to vaccine candidates. Additionally, many patients who have had viral syndromes and either tested negative for SARS-CoV-2 or were unable to receive testing may be interested in antibody testing to determine if they may have had a prior infection. Lastly, if antibodies are shown to be protective against future infection, knowing an individual’s antibody status may help with risk assessment for particular activities.
How is Antibody Testing Performed?
There are two main serological assay types used to test for SARS-CoV-2 antibodies: enzyme-linked immunosorbent assays (ELISAs) and lateral flow assays (summarized in Figure 1). While the details of the biochemistry are beyond the scope of this FLARE, both of these tests use viral antigens to detect the presence of virus-specific antibodies. The lateral flow assay (familiar to most by way of pregnancy tests) is simple to perform and can be widely, rapidly, and easily deployed - potentially even in the home. However, these assays are limited to binary outputs, as they report only the presence or absence of antibody. In comparison, ELISA offers the advantage of quantitating antibody titer, which may offer additional accuracy in determining immunity. However, ELISA testing requires more equipment, time, and expertise (Krammer and Simon 2020).
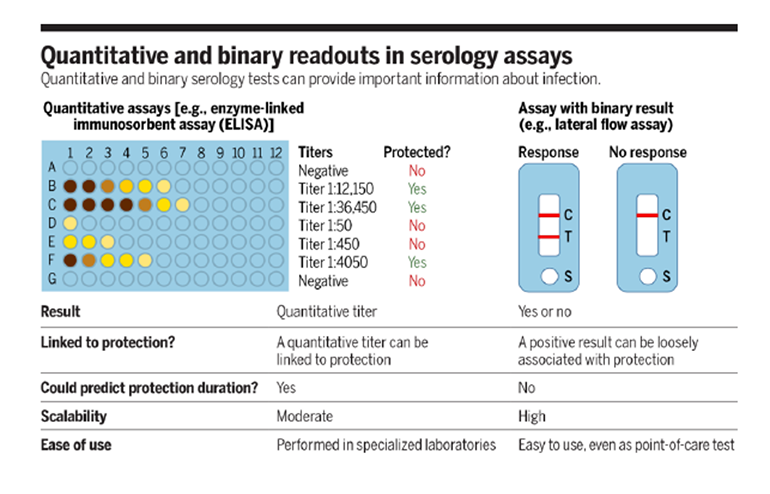
Figure 1
Comparison of ELISA and lateral flow assay for of serologic testing (Krammer and Simon 2020).
Currently available SARS-CoV-2 antibody tests have been approved under an FDA emergency use authorization (EUA). EUAs allow for rapid use of diagnostic tests without undergoing the full review and approval process, facilitating rapid testing implementation but decreasing the rigor with which new tests are evaluated. To date, FDA has evaluated 16 commercially available antibody tests: sensitivity and specificity range from 87-100% and 96-100%, respectively - this variability has important implications for interpretation of serological testing.
Antibody Response to Sars-Cov-2: Data Are Promising, but Timing Is Critical
Small initial studies have demonstrated that the majority of patients with SARS-CoV-2 infection develop antibodies against the virus (Amanat et al. 2020; Xiang et al. 2020; To et al. 2020) and several have attempted to characterize the kinetics of this seroconversion. An analysis of 29 patients revealed that antibodies were detectable as early as 4 days following confirmed infection (Figure 2). Preliminary data suggest that IgM seropositivity peaks between weeks two and three after symptom onset and starts to decline thereafter, while the IgG response lags but is longer lasting (Xiao, Gao, and Zhang 2020). The prevalence of IgM seroconversion increased significantly after the 9th day from symptom onset, with prevalence of IgG seroconversion increasing after the 11th day (Xiang et al. 2020). Another study examined 16 patients, finding 50% to harbor detectable IgG at 7 days and 100% at 14 days post-symptom onset (Wölfel et al. 2020).
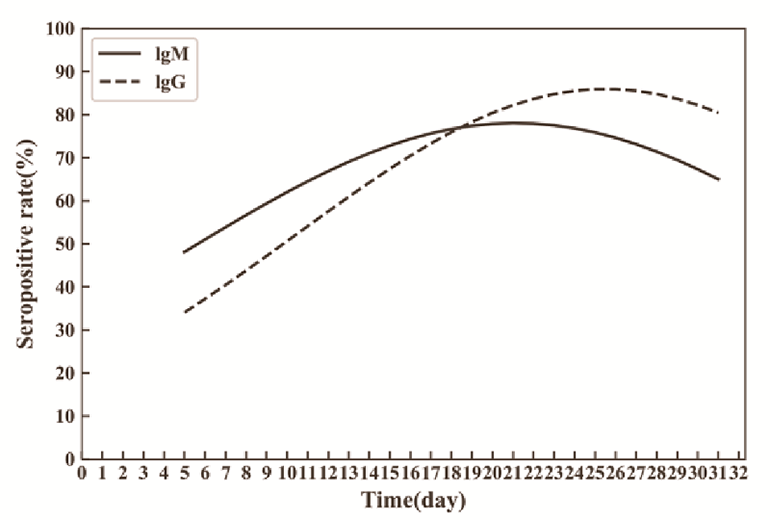
Figure 2
Changes in seropositivity prevalence over time in a cohort of patients with SARS-CoV-2 infection (Xiang et al. 2020).
A recent review provides an overview of diagnostic testing and a timeline of likely viral and antibody detection (Figure 3), highlighting the importance of considering time since likely infection in the selection and interpretation of antibody tests (Sethuraman, Jeremiah, and Ryo 2020).
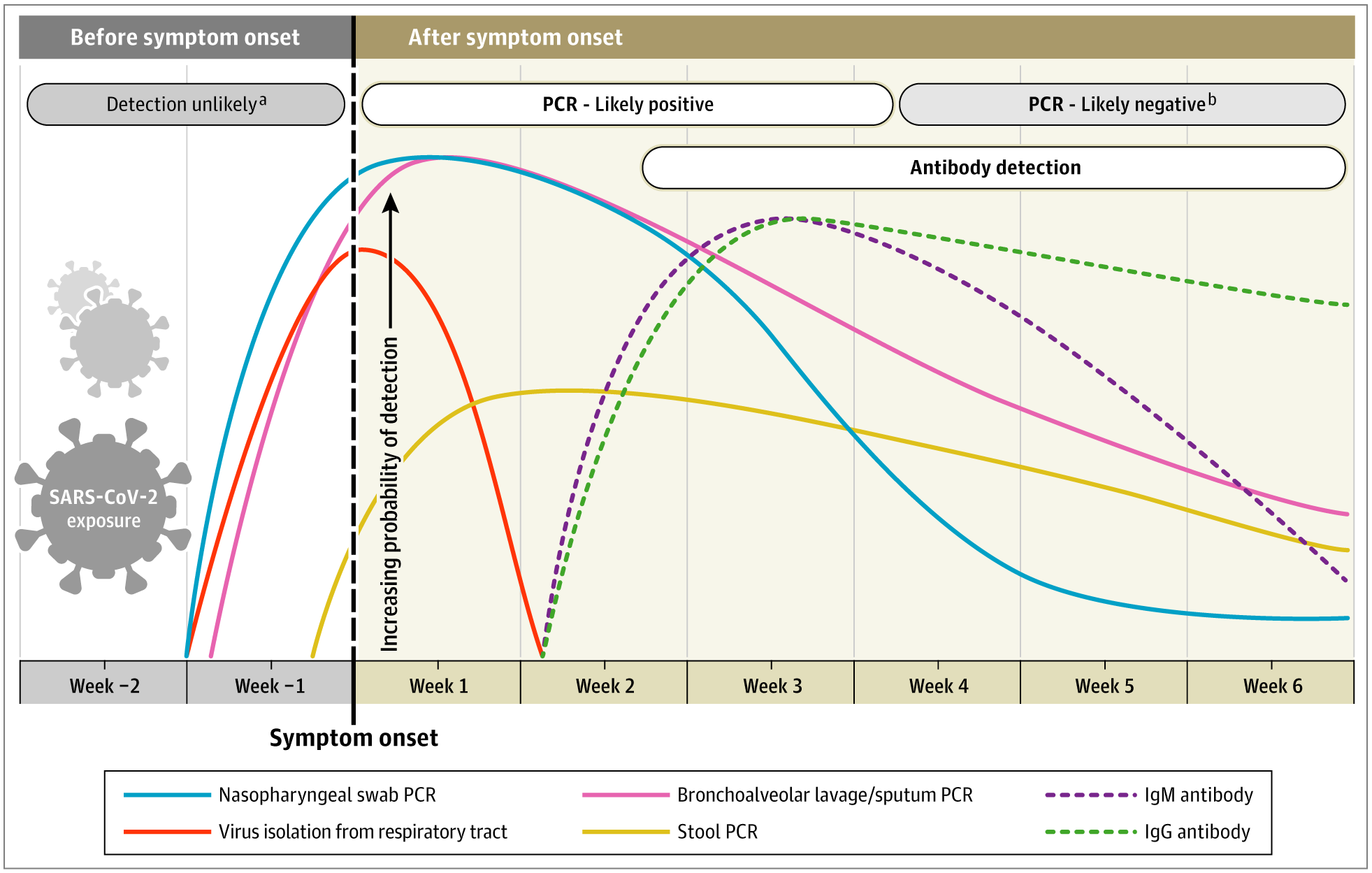
Figure 3
Estimated timing of detectable viral infection and detectable antibodies. Nasopharyngeal swab PCR (blue line) is most likely to be positive prior to symptom onset, while other viral testing including respiratory tract (orange line), BAL/sputum PCR (pink line) and stool PCR have different temporal patterns. In this figure, the authors estimate the IgM and IgG antibody response to be detectable starting in the second week after symptom onset with more durable presence (Sethuraman 2020).
Choice of Antigen Target and Risk of Cross Reactivity
One challenge inherent to antibody testing lies in selecting one or a few target epitopes from the many potential targets. This challenge is exacerbated by our current ignorance as to the precise immune correlates of protective immunity (earlier FLARE). Moreover, since essentially all immunocompetent adults harbor anti-coronavirus antibodies, selected target epitopes must be specific to SARS-CoV-2 to avoid cross reactivity and false-positives (Gorse et al. 2010).
The most common antigens used in SARS-CoV-2 antibody testing are the intact spike protein (a surface glycoprotein required for cell entry), the receptor-binding domain of the spike protein (S-RBD), and the viral nucleoprotein. Given the substantial sequence homology between the SARS-CoV-2 nucleoprotein and other coronaviruses, more cross-reactivity is predicted if the viral nucleoprotein is used (Krammer and Simon 2020).
Ni et al. examined antigenic targets of the antibody response in 8 patients with confirmed SARS-CoV-2 infection (Ni et al. 2020). Antibodies against the nucleocapsid protein and S-RBD were identified in patients but absent in healthy controls. Another study also confirmed robust antibody response against the S-RBD and the full length spike protein without a response in controls. Importantly, there did not seem to be cross reactivity among controls with antibodies to spike proteins from other coronaviruses (Amanat et al. 2020).
The association with neutralizing activity as well as the lack of cross reactivity with other coronaviruses makes the S-RBD an attractive antigen for assessing antibody response and it is commonly (though not exclusively) used in commercial tests. Understanding the risk of cross-reactivity, regardless of what specific antibodies are assessed, is essential to interpreting test results and identifying false positive results.
Clinical Performance of Antibody Testing
While it appears that most patients with SARS-CoV-2 infection develop an antibody response, it is important to understand the antibody test performance characteristics in order to correctly interpret the results.
The previously cited study by Xiang et al. also examined the utility of testing in 24 patients with clinically suspected infection but with negative RT-PCR testing. Of these patients, 21 had an IgM response and 17 had an IgG response, demonstrating potential utility in this patient population. 60 patients without clinical suspicion for SARS-CoV-2 infection revealed only 3 with IgG seroconversion and none with IgM seroconversion. Whether these rare, asymptomatic but seropositive individuals represent false positives or unrecognized infection remains to be determined (Xiang et al. 2020).
One method by which to discriminate between false positives and asymptomatic infection is to test serum collected prior to the emergence of SARS-CoV-2. Bryan and colleagues compared results of anti-SARS-CoV2-2 antibodies in 125 patients with confirmed SARS-CoV-2 infection to 1020 historical controls. Notably, every patient with confirmed disease tested positive by day 17 from symptom onset (100% sensitive) and only 1 false positive was detected in the control cohort (Bryan et al. 2020). These data indicate that in a carefully controlled setting with appropriate test selection, serologic testing for SARS-CoV-2 can have a very high sensitivity and specificity.
Points of Caution
Despite the promising data reviewed above, important concerns around antibody testing remain:
- The relationship between any one positive antibody test and protection from future SARS-CoV-2 infections (i.e. protective immunity) is unknown at this time
- Antibody testing cannot speak to infectivity and contagiousness. Individuals with SARS-CoV-2 seropositivity may nevertheless shed contagious virus and, conversely, seronegativity does not rule out acute infection
- The large number of rapidly-developed and deployed antibody tests require additional degree of caution, as test characteristics of a given assay cannot be extrapolated to others
Disease Prevalence Affects Predictive Values
Interpretation of any test result requires an understanding of the pre-test probability of disease as defined both by patient characteristics and by disease prevalence (unfortunately unknown for SARS-CoV-2). Stated differently, clinicians are most interested in the positive (PPV) and negative (NPV) predictive values of a test - parameters contingent on population prevalence. The table below highlights how dramatically PPV can vary for a test of fixed sensitivity and specificity (both 99% for the purposes of the example). Remarkably, for a disease with population prevalence 1%, a positive test result is as likely to represent a false positive as a true positive!
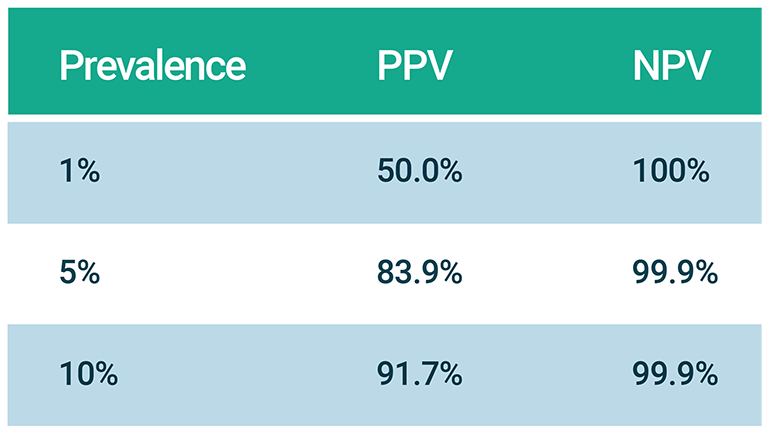
Table 1
Effect of prevalence on positive predictive value (PPV) and negative predictive value (NPV) of a test that is both 99% sensitive and specific.
So What is the Seroprevalence of SARS-CoV-2?
In short, we don’t know. As indicated above, a potential indication for antibody testing is to determine the percent of the population who have antibodies, which may be useful both in understanding the pandemic thus far and for predicting the future course of the outbreak. Although a number of studies have estimated seroprevalence, many of them have unfortunately received media attention inversely proportional to the quality of the methodology. For others, the quality of the methodology cannot be completely assessed because the authors have shared their results in media reports rather than scientific papers. Few large-scale studies exist and estimated prevalence ranges widely from 2% to over 30%.
The highest reported prevalence comes from a study of 200 people in Chelsea, MA. As covered in a prior FLARE (April 21), Chelsea has been hard hit by COVID-19. Researchers from MGH tested blood from 200 volunteers and found that 31% had antibodies. This research has not yet been published (it has been reported locally and was described by the Boston Globe) so it is not possible to thoroughly evaluate the methodology. The study has been criticized both for recruiting a non-random sample and for using a test with a specificity as low as 90%, potentially resulting in a high false positive rate.
Sampling biases may have confounded other widely-publicized seroprevalence studies as well. A study from Germany (to date only available as a preprint) reported a roughly 15% prevalence, sampling 919 individuals from 405 households (Streeck et al. 2020). These are clearly not independent samples - members of the same household are highly likely to have concordant antibody status. The lack of generalizability did not stop the authors from conducting extensive media interviews. Similar concerns have been expressed about a seroprevalence study (Bendavid et al. 2020) conducted in Santa Clara County, CA in which the authors reported a seroprevalence rate of 1.5%. They subsequently adjusted this for demographic and test performance criteria (sensitivity 83% and specificity 99%) to 2.8%. The study recruited subjects via Facebook ads and so it likely also suffers from selection bias. So far this study is also available only as a preprint. In another similarity to the German study, the methodologic shortcomings and lack of peer review have not stopped the authors from conducting extensive media tours.
Some of the same authors (Sood et al. 2020) attempted to address the flaws of their Santa Clara study by conducting one of the few studies on seroprevalence that has been subject to peer review. They used a test with an estimated sensitivity of 83% and a specificity of 99%. They used a randomly generated sample of residents in Los Angeles County, but only approximately 50% of invited participants agreed to be tested. The tested group failed to match county demographics and the proportion of positive tests did in fact vary by ethnic group. In addition, ~13% of their sample was symptomatic, likely indicating that symptomatic people were more likely to participate.
The largest seroprevalence study to date was conducted in Boise, Idaho (Bryan et al. 2020). These authors first carefully determined the specificity of their assay by testing a large number of serum samples predating SARS-CoV-2 and found only one false positive from 1020 samples, suggesting a specificity of ~99%. They then tested serum from patients with PCR confirmed SARS-CoV-2 infection and found a sensitivity of roughly 50% on day 7 from symptom onset, which rose to 100% by day 17 from symptom onset. They then tested 4,856 community members as part of an initiative to determine the community wide prevalence. While this is a large number of individuals, they were self-selected. The authors report a prevalence of 1.79% which should likely be regarded as something of an overestimate given sampling bias.
Other studies attempt to analyze the population prevalence of disease not with antibody testing, but by using various modeling techniques to extrapolate from hospitalization data. This was the approach used by Ceylan and colleagues to arrive at an estimated prevalence of 4.4% for the nation of France (Ceylan 2020). Unfortunately, the relationship between hospitalized cases and prevalence of mild or asymptomatic infection was based on data from the Diamond Princess cruise ship outbreak, in which all passengers were tested. It is unclear whether data from this confined population is generalizable to the national level.
In sum, the available data on seroprevalence are of low quality and very few studies have been subjected to peer review. In our opinion, none are of high enough quality to drive evidence-based decision making. Given the intense public interest in this issue, it seems wisest for researchers to avoid speaking to the media prior to formally vetting their results. The uncertainty about the quality of different antibody tests (discussed above) adds yet another level of uncertainty about the prevalence of SARS-CoV-2.
Conclusions
SARS-CoV-2 serologic assays are a critical tool for diagnosing prior infection and assessing antibody seroprevalence within a population. There are numerous serologic assays, but limitations exist that can lead to both false negatives and false positives. Serologic testing will be critical to define the immune correlates of protective immunity to SARS-CoV-2 and may be of great use in understanding the dynamics of the pandemic. Consequently, we must take advantage of newly available, high quality serologic tests in a thoughtful and strategic manner to improve our understanding of and response to the COVID-19 pandemic.
References:
- Amanat, Fatima, Daniel Stadlbauer, Shirin Strohmeier, Thi Nguyen, Veronika Chromikova, Meagan McMahon, Kaijun Jiang, et al. 2020. “A Serological Assay to Detect SARS-CoV-2 Seroconversion in Humans.” Allergy and Immunology. medRxiv. https://doi.org/10.1101/2020.03.17.20037713.
- Bendavid, Eran, Bianca Mulaney, Neeraj Sood, Soleil Shah, Emilia Ling, Rebecca Bromley-Dulfano, Cara Lai, et al. 2020. “COVID-19 Antibody Seroprevalence in Santa Clara County, California.” Epidemiology. medRxiv. https://doi.org/10.1101/2020.04.14.20062463.
- Bryan, Andrew, Gregory Pepper, Mark H. Wener, Susan L. Fink, Chihiro Morishima, Anu Chaudhary, Keith R. Jerome, Patrick C. Mathias, and Alexander L. Greninger. 2020. “Performance Characteristics of the Abbott Architect SARS-CoV-2 IgG Assay and Seroprevalence in Boise, Idaho.” Journal of Clinical Microbiology. https://doi.org/10.1128/jcm.00941-20.
- Ceylan, Zeynep. 2020. “Estimation of COVID-19 Prevalence in Italy, Spain, and France.” The Science of the Total Environment 729 (April): 138817.
- Gorse, Geoffrey J., Gira B. Patel, Joseph N. Vitale, and Theresa Z. O’Connor. 2010. “Prevalence of Antibodies to Four Human Coronaviruses Is Lower in Nasal Secretions than in Serum.” Clinical and Vaccine Immunology: CVI 17 (12): 1875–80.
- Krammer, Florian, and Viviana Simon. 2020. “Serology Assays to Manage COVID-19.” Science, May. https://doi.org/10.1126/science.abc1227.
- NIAID. n.d. “SARS-COV2 Pandemic Serosurvey and Blood Sampling.” Accessed May 22, 2020. https://www.niaid.nih.gov/clinical-trials/sars-cov2-pandemic-serosurvey-blood-sampling.
- Ni, Ling, Fang Ye, Meng-Li Cheng, Yu Feng, Yong-Qiang Deng, Hui Zhao, Peng Wei, et al. 2020. “Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals.” Immunity, May. https://doi.org/10.1016/j.immuni.2020.04.023.
- Sethuraman, Nandini, Sundararaj Stanleyraj Jeremiah, and Akihide Ryo. 2020. “Interpreting Diagnostic Tests for SARS-CoV-2.” JAMA: The Journal of the American Medical Association, May. https://doi.org/10.1001/jama.2020.8259.
- Sood, Neeraj, Paul Simon, Peggy Ebner, Daniel Eichner, Jeffrey Reynolds, Eran Bendavid, and Jay Bhattacharya. 2020. “Seroprevalence of SARS-CoV-2-Specific Antibodies Among Adults in Los Angeles County, California, on April 10-11, 2020.” JAMA: The Journal of the American Medical Association, May. https://doi.org/10.1001/jama.2020.8279.
- Streeck, Hendrik, Bianca Schulte, Beate Kuemmerer, Enrico Richter, Tobias Höller, Christine Fuhrmann, Eva Bartok, et al. 2020. “Infection Fatality Rate of SARS-CoV-2 Infection in a German Community with a Super-Spreading Event.” medRxiv.
- To, Kelvin Kai-Wang, Owen Tak-Yin Tsang, Wai-Shing Leung, Anthony Raymond Tam, Tak-Chiu Wu, David Christopher Lung, Cyril Chik-Yan Yip, et al. 2020. “Temporal Profiles of Viral Load in Posterior Oropharyngeal Saliva Samples and Serum Antibody Responses during Infection by SARS-CoV-2: An Observational Cohort Study.” The Lancet Infectious Diseases. https://doi.org/10.1016/s1473-3099(20)30196-1.
- Wölfel, Roman, Victor M. Corman, Wolfgang Guggemos, Michael Seilmaier, Sabine Zange, Marcel A. Müller, Daniela Niemeyer, et al. 2020. “Virological Assessment of Hospitalized Patients with COVID-2019.” Nature, April. https://doi.org/10.1038/s41586-020-2196-x.
- Xiang, Fei, Xiaorong Wang, Xinliang He, Zhenghong Peng, Bohan Yang, Jianchu Zhang, Qiong Zhou, et al. 2020. “Antibody Detection and Dynamic Characteristics in Patients with COVID-19.” Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America, April. https://doi.org/10.1093/cid/ciaa461.
- Xiao, Ai Tang, Chun Gao, and Sheng Zhang. 2020. “Profile of Specific Antibodies to SARS-CoV-2: The First Report.” The Journal of Infection, March. https://doi.org/10.1016/j.jinf.2020.03.012.
View all COVID-19 Updates
Learn more about research in the Division of Pulmonary and Critical Care Medicine