Not All Recurrent Mutations Are Cancer Drivers
Key findings
- This study investigated how mesoscale genomic features—those affecting dozens of base pairs but not vast stretches of the genome—affect mutational recurrence in human tumors
- Common mesoscale features known as DNA "hairpins" were associated with recurrence of mutations outside of known cancer drivers
- This finding challenges the presumption that recurrent mutations must be drivers and highlights the importance of incorporating mesoscale features into the analysis of cancer genomes
Subscribe to the latest updates from Oncology Advances in Motion
A central assumption in cancer genome research is that mutational processes are random. Thus, if the same DNA base pair is mutated recurrently across patients, this "hotspot" mutation must be contributing to tumor development.
Michael S. Lawrence, PhD, and Lee Zou, PhD, of the Massachusetts General Hospital Cancer Center, and colleagues have called this tenet into question. In Science, they report that some recurrent tumor mutations are apt to be unimportant to cancer progression—and others may be novel cancer drivers.
Meeting in the Middle
Most cancer genome analyses examine mutations on a small scale (one to three base pairs) or the megabase level. Little attention has been paid to the mesoscale (~30 base pairs), which includes common features such as DNA "hairpins," also known as stem-loops. Formation of these loops can "flip out" bases, increasing their exposure to endogenous mutation.
The researchers examined the distribution of mutations in more than 9,000 human tumors. They found that APOBEC3A, a cytidine deaminase enzyme, has a substantial preference for attacking DNA hairpins, resulting in recurrent mutations.
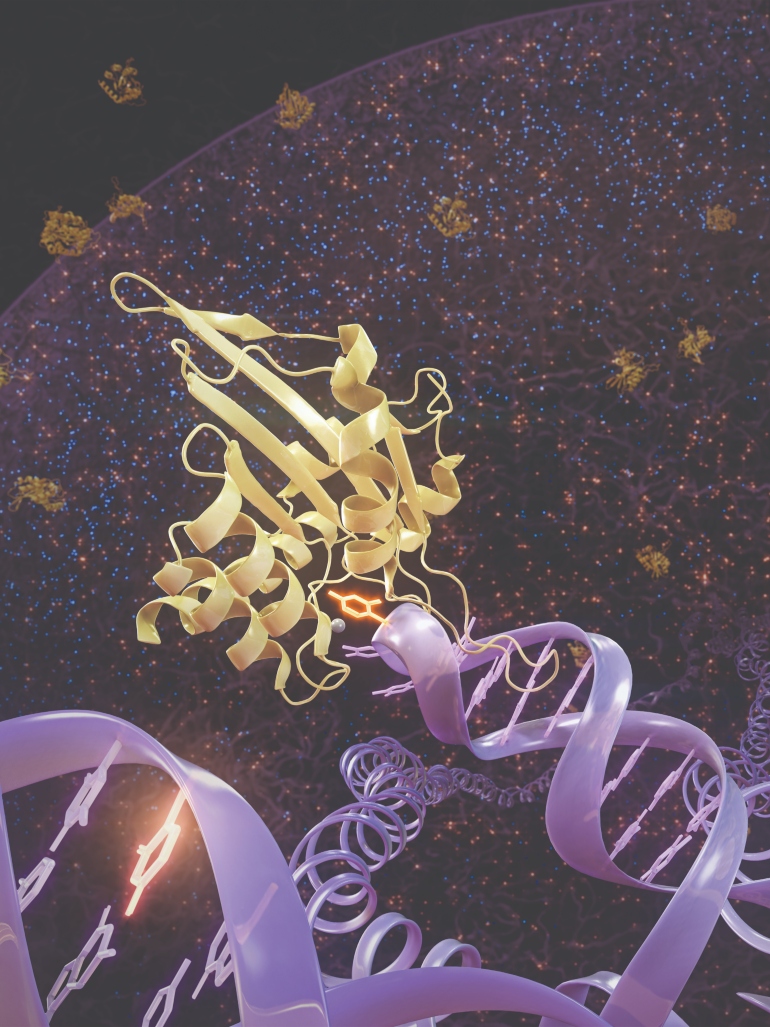
The mutational landscape of a cancer cell across size regimes. At the smallest scale, local DNA trinucleotide sequences (lower-left foreground) correlate with the "mutational signatures" induced by various mutagens. At the largest scale (background of image), chromatin is organized into multi-megabase domains comprising Compartment B (tightly packed, gene-poor DNA lining the nuclear periphery) and Compartment A (gene-rich open DNA in the nuclear interior). Mutations induced by APOBEC enzymes (yellow points) are distributed equally across the two compartments, but most other types of mutations (blue points) are concentrated in Compartment B. Between the large and small extremes lies the "mesoscale" regime, where genomic features like hairpin-forming ability are determined. DNA exposed in a hairpin loop is vulnerable to attack by the enzyme APOBEC3A (center), giving rise to highly recurrent passenger mutations in cancer. Credits: Lawrence and Zou Laboratories (MGH), artistic: www.SciStories.com.
Two Types of Mutation Hotspots
After analyzing the preferred substrates of APOBEC3A, the researchers examined whole-exome sequencing data on 2,572 APOBEC+ human tumors. Of the top 100 most frequently mutated sites:
- 45 comprised recurrent mutations at preferred APOBEC3A sites, but in genes not known to be associated with cancer
- 55 were in genes previously described as cancer drivers (e.g., PIK3CA, TP53), but all except one occurred at non-optimal APOBEC3A sites
Thus, some recurrent mutations may be "passenger hotspots," contributing no advantage to cancer cells but observed at high frequency because of the ease with which they are generated. On the other hand, some recurrent non-hairpin APOBEC mutations may be novel cancer drivers. The ability to discriminate between passenger and driver hotspots will be essential to developing novel cancer therapies.
view original journal article Subscription may be required
Learn more about research at the Mass General Cancer Center
Refer a patient to the Mass General Cancer Center