Novel Noninvasive Scoring System Predicts Presence of Obstructive CAD
Key findings
- Urgent public health need exists for a non-invasive, cost-effective scoring system to predict CAD risk
- New predictive scoring system created by first identifying and then adding biomarker and clinical variables
- The system combines two clinical characteristics with four biomarker concentration levels: midkine, adiponectin, apo C-I and KIM-1
Coronary artery disease (CAD) is a leading cause of suffering, disability and premature death worldwide. Patients could benefit and public health could improve from the development of a non-invasive scoring system that predicts the presence of anatomically significant CAD.
Subscribe to the latest updates from Cardiovascular Advances in Motion
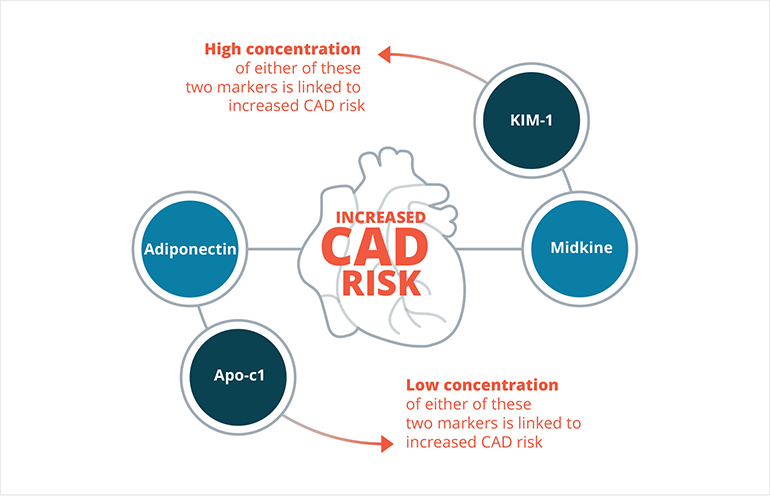
Fig. 1: The CAD Scoring System’s Four Biomarkers Were Proven to Correlate CAD Risk Based on Either High or Low Levels of Concentration
High diagnostic accuracy
In an international collaboration, researchers from Massachusetts General Hospital and Harvard Clinical Research Institute identified clinical and biomarker predictors of clinically-significant CAD to develop such a diagnostic scoring system. They hypothesized that adding plasma biomarkers to known clinical risk factors might increase predictive accuracy.
By reviewing more than 200 candidate variables, 109 of which were biomarkers, investigators identified predictors of ≥ 70% stenosis in at least one major coronary vessel. Based on this analysis, they combined two clinically-relevant variables, male sex and previous percutaneous coronary intervention, with four biomarkers: midkine, adiponectin, apo C-I and KIM-1 from blood samples of patients enrolled in the CASABLANCA study.
Preliminary results showed high accuracy in predicting obstructive CAD, defined as ≥70% stenosis. At optimal cutoff the score had 77% sensitivity, 84% specificity and a positive predictive value of 90% for ≥ 70% stenosis.
Two low-concentration biomarkers
Patients with severe CAD had lower levels of two biomarkers: adiponectin and apo C-I.
Adiponectin is an amino acid peptide secreted by adipose tissue that helps regulate glucose and fatty acid metabolism. Previous studies have linked low-levels of adiponectin with CAD and the progression of vascular disease.
Apo C-I is a key regulator of triglycerides in both fasting and postprandial conditions and has been previously identified as a predictor of CAD, although there is conflicting data from clinical studies.
Two high-concentration biomarkers
Patients with severe CAD also had higher levels of two other biomarkers: KIM-1 and midkine.
KIM-1 is a proximal renal tubular marker whose concentration levels have been linked to acute kidney injury. The current study is believed to be the first to associate KIM-1 levels with the presence of obstructive CAD.
Midkine is a cytokine/growth factor found in multiple organs and implicated in the pathophysiology of CAD through multiple pathways such as plaque infiltration of inflammatory cells. The current study appears to be the first to implicate a diagnostic role for midkine levels in CAD in humans.
Further testing of the scoring system is underway to validate these results and demonstrate broad clinical utility.
view original journal article Subscription may be required
Refer a patient to the Massachusetts General Hospital Corrigan Minehan Heart Center
