Pathways Case Report: A Case of Unusually High Penetrance of a Classic Thrombophilia Mutation
In This Case Study
- A 65-year-old woman with known heterozygous prothrombin G20210A mutation (PGM) with a family history of stroke presented to the hospital with shortness of breath and chest pain despite taking prophylactic anticoagulation therapy
- One month prior to presentation, she developed acute difficulty swallowing and right-sided neurologic deficits. She was diagnosed with an intraparenchymal brain bleed that required a craniotomy
- She was transferred to Massachusetts General Hospital for anticoagulation management
A 65-year-old woman with known heterozygous prothrombin G20210A mutation (PGM) with a family history of stroke presented to the hospital with shortness of breath and chest pain despite taking prophylactic anticoagulation therapy. One month prior to presentation, she developed acute difficulty swallowing and right-sided neurologic deficits. She was diagnosed with an intraparenchymal brain bleed that required a craniotomy. She was discharged to a rehabilitation facility given her limited mobility. While at this facility, she developed difficulty breathing and chest pain. Initial evaluation ruled out a myocardial infarction. The patient demonstrated persistent symptoms, and a CT pulmonary angiogram revealed multiple occlusive and nonocclusive pulmonary emboli. In addition, an ultrasound showed a deep venous thrombosis on her right lower extremity. She was transferred to Massachusetts General Hospital for anticoagulation management.
Subscribe to the latest updates from Neuroscience Advances in Motion
Upon admission, the patient was normotensive with normal oxygen saturation levels. Workup was consistent with low-risk pulmonary emboli, as revealed by unremarkable aminoterminal pro-B-type natriuretic peptide (NTproBNP) and troponin blood tests. An echocardiogram showed no evidence of right heart strain. Neurosurgery, hematology, and pulmonology were consulted, and she was started on low-dose heparin with serial CT head imaging and neurologic examinations. After intraparenchymal hemorrhage stability was confirmed, she was transitioned to the anticoagulant apixaban twice daily. She was discharged with plans to continue anticoagulation for three months. Her venous thromboembolism was in the setting of immobility and critical illness.
The Pathways Service in the Department of Medicine at Massachusetts General Hospital was consulted and focused on prothrombin mutations in disease.
Background and Diagnosis
Prothrombin (Factor II) is the inactive precursor protein of thrombin (Factor IIa), the final enzyme in the clotting cascade. It is made in the liver and is converted to thrombin by the prothrombin activator complex (composed of Factor Xa, Factor V, phospholipid, and calcium ions). Thrombin facilitates clot formation by promoting fibrinogen conversion to fibrin, activating Factor XIII for clot stabilization, and inhibiting fibrinolysis via activation of the thrombin-activatable fibrinolysis inhibitor (TAFI). Increased levels of prothrombin lead to greater clot formation and stabilization.
DNA sequence changes, such as the prothrombin G20210A and Factor V Leiden gene mutation, lead to changes in thrombin conversion and increased risk of clotting. Notable to our case, G20210A mutations are the second most common thrombophilia with an autosomal dominant inheritance pattern and incidence of 2-5%, which occurs with greater incidence in the white population. Despite its high incidence in the population, it has variable penetrance as shown by the lack of venous thromboembolism (VTE) development in most individuals carrying the mutation. Our patient's family history of thrombophilia was notable for its high penetrance of clotting events. Of the eight siblings, five have the prothrombin G20210A mutation, and of those, three have had a VTE. This significantly differs from the expected incidence of clots for patients with this mutation (~10%).
The prothrombin G20210A mutation is located at or near the cleavage and polyadenylation site of the prothrombin mRNA and increases prothrombin expression by 1.2 to 1.4 fold. Patient samples and reporter constructs revealed that the G20210A mutation does not impact mRNA stability but does affect polyadenylation site usage1, 2. The polyadenylation site is heterogeneous, and studies suggest that G20210A mutation causes a preference for mRNA polyadenylation at a different site than wildtype (20210, rather than 20212). Usage of this site causes increased expression3. Altering the polyadenylation site results in a changed secondary structure and increased binding by an RNA regulatory protein4. Taken together, these studies suggest that mutation of the nucleotide at 20210 causes a change in cleavage and polyadenylation site, which in turn affects RNA secondary structure and modulates binding of regulatory factors that control protein translation.
The prothrombin G20210A mutation occurs in the 3'UTR of the gene rather than the coding region5. Several binding and regulatory elements within the 3'UTR (three prime untranslated region) control the stability, translation, and localization of mRNA transcripts. Furthermore, this region is regulated through [1] iron-response elements which can stabilize mRNA transcripts through the binding of iron regulatory proteins; [2] microRNA response elements (miRNA binding sites) which can cause translational repression of mRNA molecules when bound; [3] AU rich-elements (AREs) which are targets for AU rich element binding proteins and can serve both destabilizing (in the case of many cytokines) and stabilizing roles; [4] cytoplasmic polyadenylation elements which can increase polyadenylation and translational efficiency through binding of polyadenine binding proteins; and [5] proteins which aid in secondary structural looping and therefore allow mRNA to form a more accessible or less accessible structure for translation (Figure 1).

Figure 1
Regulatory elements in the 3' untranslated region of mRNA. Detailed regions highlight IRE (iron-response elements), IRP (Iron-regulatory proteins), miRNAs (bound to MicroRNA response elements), AU rich-elements, Cytoplasmic polyadenylation elements (CPE), and the terminal polyadenylation sites on mRNA.
The 3'UTR region is vital in the regulation of mRNA of several oncogenes. Indeed, as miRNA molecules play an important role in the suppression of gene expression by binding to the 3'UTR, loss of this region can drive activation and maintenance of oncogenes6. The miRNAs and AREs regulatory regions are associated with disease phenotypes. Two interesting modulations related to miRNAs in the genetic code of the 3'UTR include rs13702 in lipoprotein lipase, which is associated with HDL-cholesterol levels, and rs12190287 in transcription factor 21 (TCF21), which is associated with coronary artery disease7. Further mutations in the AREs of the 3'UTR of bone morphogenic protein 2 (BMP2), cyclooxygenase-2 (COX2), and in beta-2 adrenergic receptor (B2-AR) are associated with osteoporosis, colorectal cancer, and variable response to B2 agonists, respectively8.
In addition to the remarkable presentation of mutations in our patient's family, there were also several members, including two with prothrombin gene mutation and early-onset Parkinson's disease (PD). The genetics of PD is multifactorial with over 100 distinct genetic loci associated with the disease. As our patient's family had high incidences of both the prothrombin G20210A gene mutation as well as PD, we considered the possibility that there may be a relationship between increased prothrombin levels and development of PD. We performed an initial evaluation of the possible relationship between the prothrombin G20210A mutation and the development of PD utilizing the UK Biobank, a freely available navigation tool built by Ben Neale's laboratory at Massachusetts General Hospital. Although initial evaluation did not reveal a strong correlation with PD, we did find some association between this mutation and patients who were prescribed pramipexole (a medication used in the treatment of PD) (Figure 2). Interestingly, recent work describes how the Kringle-2 domain of prothrombin, once made available to the bloodstream after cleavage by factor X, can inappropriately activate microglia within the brain and lead to neuronal cell death (ultimately leading to neurocognitive issues)9. Additionally, an increase in levels of the Kringle-2 domain has been observed in the brains of a murine model of Alzheimer's disease. Intriguingly, these Alzheimer's disease mice were partially protected from having neurocognitive effects when they were given the factor 10 inhibitor, Rivaroxaban10.
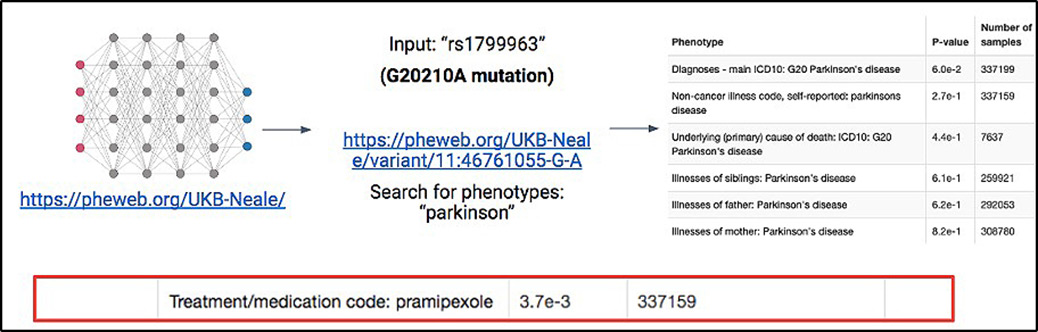
Figure 2
Association between G20210A prothrombin mutation and Parkinson's disease. Experimental design to analyze the G20210A mutation within the UK Biobank online resource and the possible relationship between these individuals and Parkinson's disease.
Summary and Future Steps
Our patient's presentation was remarkable for two reasons. First, although the prothrombin gene mutation has variable penetrance across the population, nearly all the carriers in her family have exhibited clotting phenotypes. Second, her family history was also notable for several members, including two with prothrombin gene mutation, with early-onset Parkinson's disease.
To test our first hypothesis that our patient possesses additional genetic risk factors for thrombosis, we recommend the following five steps: [1] Expanded genetic testing for known polymorphisms significant in thrombosis (use the extended thrombophilia panel containing 55 novel mutations associated with increased risk of VTE11); [2] Whole genome/exome sequencing of the patient, siblings, and parents; [3] Analysis of co-occurring mutations in large datasets such as resources available through the UK Biobank to identify polymorphisms that, together with prothrombin gene mutation, confer an increased risk of clotting phenotypes; [4] Genetic screen to identify factors important to prothrombin by using previously developed reporter constructs of the 3' UTR of prothrombin3 to transfect liver cells and perform a whole genome screen using a CRISPR library. Genes that affect the ratio of expression between reporter constructs would be followed up for contribution to differential prothrombin levels (Figure 3); [5] Biochemical screens to identify binding partners of prothrombin 3'UTR variants (RNA pulldown experiment to identify proteins with differential binding to prothrombin 3' UTRs of interest to identify potential regulatory pathways). These experiments would provide a better understanding of risk factors in individuals harboring mutations.

Figure 3
Experimental pathway detailing proposed CRISPR screen to identify modulators of G20210A prothrombin expression.
To further evaluate the possible relationship between PD and increased prothrombin through the G20210A mutation, we propose examining available genomic data set and, in particular, analyzing the subset of prothrombin G20210A patients to those that developed VTEs and seeing if these are more closely associated with PD as these patients likely have higher levels of prothrombin. Furthermore, we could overexpress prothrombin in a murine model and evaluate the development of neurocognitive disorders in this setting. In summary, understanding the crosstalk between PD and prothrombin may lead to significant clinical benefit to patients.
References
- Pollak ES, Lam HS, Russell JE. The G20210A mutation does not affect the stability of prothrombin mRNA in vivo. Blood. 2002;100(1):359-62. doi: 10.1182/blood-2002-02-0412. PubMed PMID: 12070052.
- Liu X, Russell JE. Cytoplasmic stabilities of 3'UTR-polymorphic prothrombin mRNAs. J Thromb Haemost. 2010;8(11):2580-3. doi: 10.1111/j.1538-7836.2010.04026.x. PubMed PMID: 20723024; PMCID: PMC2998548.
- Ceelie H, Spaargaren-van Riel CC, Bertina RM, Vos HL. G20210A is a functional mutation in the prothrombin gene; effect on protein levels and 3'-end formation. J Thromb Haemost. 2004;2(1):119-27. doi: 10.1111/j.1538-7836.2003.00493.x. PubMed PMID: 14717975.
- Liu X, Jiang Y, Russell JE. A potential regulatory role for mRNA secondary structures within the prothrombin 3'UTR. Thromb Res. 2010;126(2):130-6. doi: 10.1016/j.thromres.2010.04.010. PubMed PMID: 20553951; PMCID: PMC2910803.
- Poort SR, Rosendaal FR, Reitsma PH, Bertina RM. A common genetic variation in the 3'-untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood. 1996;88(10):3698-703. PubMed PMID: 8916933.
- Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673-84. doi: 10.1016/j.cell.2009.06.016. PubMed PMID: 19703394; PMCID: PMC2819821.
- Griesemer D, Xue JR, Reilly SK, Ulirsch JC, Kukreja K, Davis JR, Kanai M, Yang DK, Butts JC, Guney MH, Luban J, Montgomery SB, Finucane HK, Novina CD, Tewhey R, Sabeti PC. Genome-wide functional screen of 3'UTR variants uncovers causal variants for human disease and evolution. Cell. 2021;184(20):5247-60.e19. Epub 20210916. doi: 10.1016/j.cell.2021.08.025. PubMed PMID: 34534445; PMCID: PMC8487971.
- Hitti E, Khabar KS. Sequence variations affecting AU-rich element function and disease. Front Biosci (Landmark Ed). 2012;17(5):1846-60. Epub 20120101. doi: 10.2741/4023. PubMed PMID: 22201840.
- Kim S, Sharma C, Jung UJ, Kim SR. Pathophysiological Role of Microglial Activation Induced by Blood-Borne Proteins in Alzheimer's Disease. Biomedicines. 2023;11(5). Epub 20230507. doi: 10.3390/biomedicines11051383. PubMed PMID: 37239054; PMCID: PMC10216844.
- Kim S, Moon GJ, Kim HJ, Kim DG, Kim J, Nam Y, Sharma C, Leem E, Lee S, Kim KS, Ha CM, McLean C, Jin BK, Shin WH, Kim DW, Oh YS, Hong CW, Kim SR. Control of hippocampal prothrombin kringle-2 (pKr-2) expression reduces neurotoxic symptoms in five familial Alzheimer's disease mice. Br J Pharmacol. 2022;179(5):998-1016. Epub 20211024. doi: 10.1111/bph.15681. PubMed PMID: 34524687; PMCID: PMC9298060.
- Lee EJ, Dykas DJ, Leavitt AD, Camire RM, Ebberink E, García de Frutos P, Gnanasambandan K, Gu SX, Huntington JA, Lentz SR, Mertens K, Parish CR, Rezaie AR, Sayeski PP, Cromwell C, Bar N, Halene S, Neparidze N, Parker TL, Burns AJ, Dumont A, Yao X, Chaar CIO, Connors JM, Bale AE, Lee AI. Whole-exome sequencing in evaluation of patients with venous thromboembolism. Blood Adv. 2017;1(16):1224-37. Epub 20170629. doi: 10.1182/bloodadvances.2017005249. PubMed PMID: 29296762; PMCID: PMC5728544.