‘Human Knockout Project’ in a Pakistani Population with High Levels of Consanguinity
In This Article
- In this poster presented at Clinical Research Day at Mass General, this researcher study leverages naturally-occurring null mutations and consanguinity in the human population to identify “human knockouts” and characterize the consequences of complete gene disruption
- The researchers sequenced the protein-coding regions of 10,503 adults living in Pakistan as part of the PROMIS: Pakistan Risk of Myocardial Infarction Study
- The researchers found that homozygous disruption of 1,317 genes is tolerated in 10,503 humans, and for several, they establish phenotypic consequences
- For one gene
(APOC3) , an emerging cardiovascular disease therapeutic target, the researchers recruited individuals with lifelong deficiency and showed that dietary absorption of fat was blunted in this setting
Subscribe to the latest updates from Cardiovascular Advances in Motion
Background
Experimental “knockout” model organisms have proven useful for understanding gene function. In this study, the researchers led by Pradeep Natarajan, MD, MMSc, director of Preventive Cardiology, and Sekar Kathiresan, MD, director of the Center for Genomic Medicine at Massachusetts General Hospital, leverage naturally-occurring null mutations and consanguinity in the human population to:
- Identify “human knockouts”
- Characterize the consequences of complete gene disruption
Methods
- The researchers sequenced the protein-coding regions of 10,503 adults living in Pakistan as part of the PROMIS: Pakistan Risk of Myocardial Infarction Study, where consanguinity rates are amongst the highest in the world, to find participants knocked out for genes
- All participants were phenotyped for >200 cardiometabolic traits and the researchers tested whether knockouts differed from wild-type across all phenotypes
- For a single gene, they performed genotype-based recall of homozygous nulls and wild-type participants followed by provocative physiologic testing
- Based on observed consanguinity, the researchers simulated the number of expected “human knockouts” in PROMIS compared to outbred Exome Aggregation Consortium populations
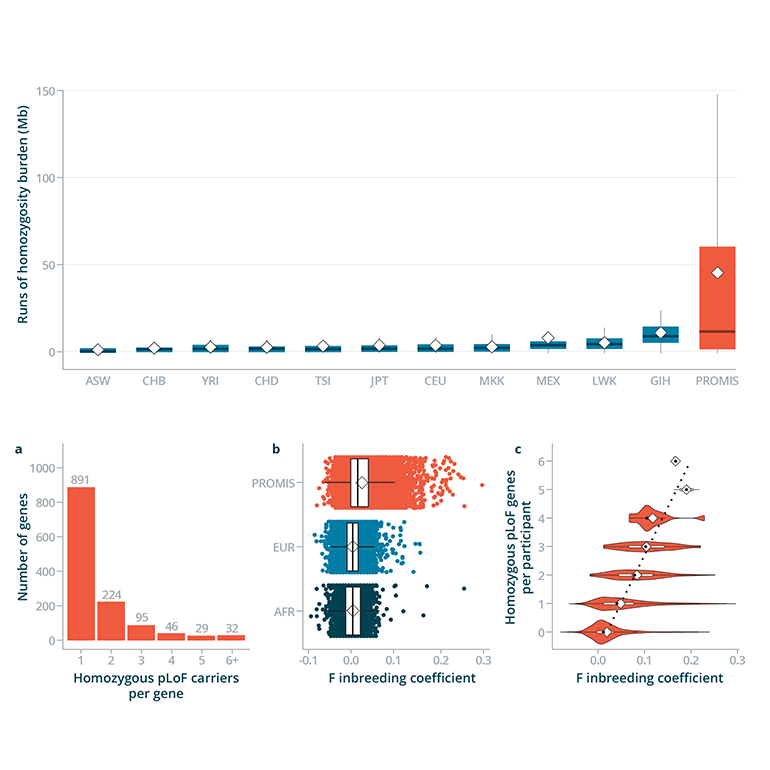
Fig. 1: Data Showing Multi-Generational Consanguinity and Burden of Homozygosity
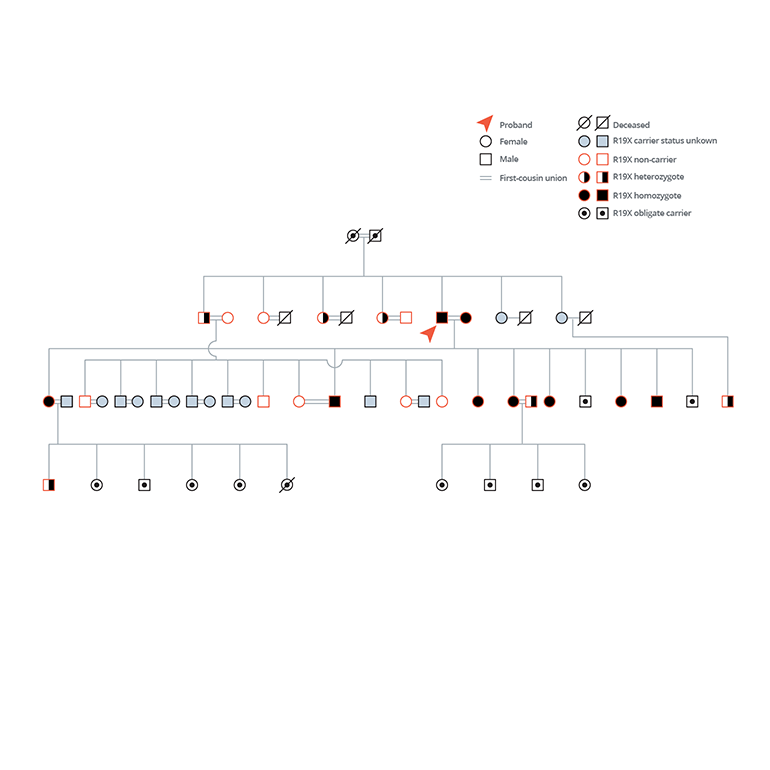
Fig. 2: Family-based Recruitment Enriches for APOC3 Knockouts
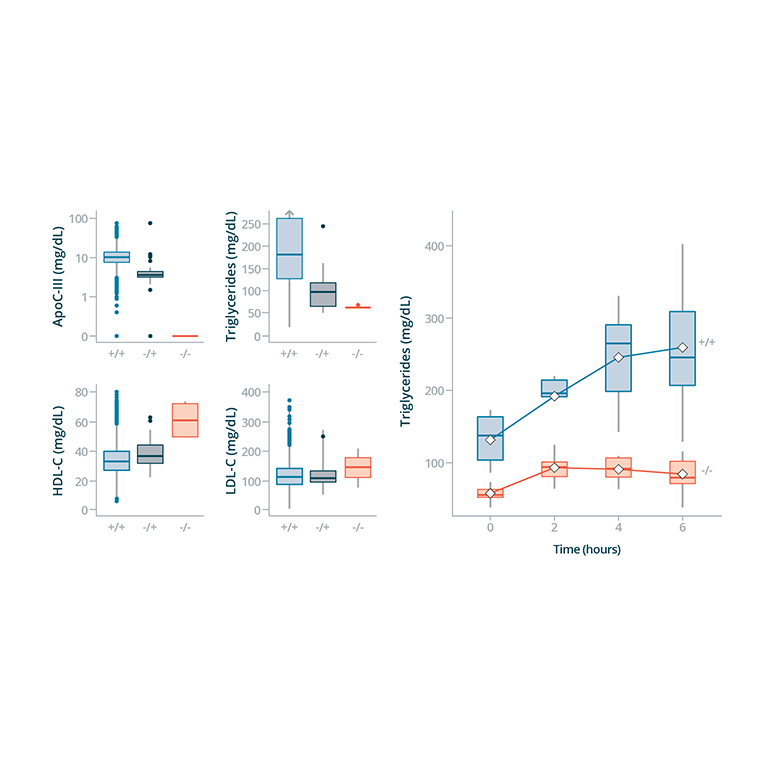
Fig. 3: Static and Dynamic Lipid Assessment by APOC3 Knockout Status
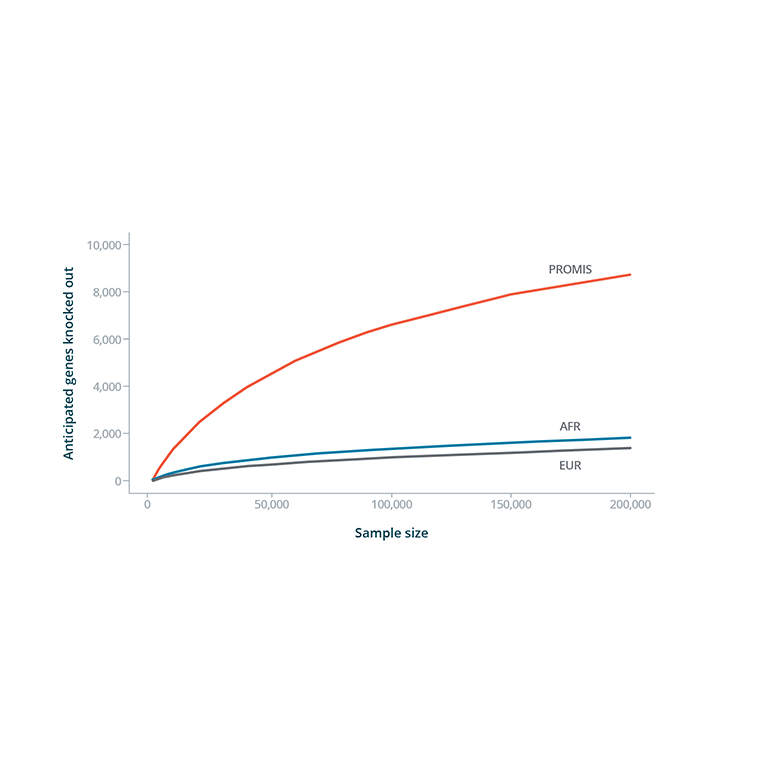
Fig. 4: Data Showing a Higher Number of Anticipated Genes Knocked Out in PROMIS Study
Conclusions
- The researchers found that homozygous disruption of 1,317 genes is tolerated in 10,503 humans, and for several, they establish phenotypic consequences
- This work lays the foundation for a comprehensive project to understand gene function by studying human knockouts
- This study was published in Nature
Authors
Pradeep Natarajan1,2, Danish Saleheen3,4, Irina A. Armean2,5, Wei Zhao3, Hong-Hee Won1,2, Sumeet Khetarpal6, Eric S Lander2, Stacey Gabriel2, Mark J Daly2,5, Philippe Frossard4, John Danesh7, Daniel J Rader6,8, Sekar Kathiresan1,2; on behalf of the PROMIS investigators
1Center for Genomic Medicine and Cardiovascular Research Center, Massachusetts General Hospital, Boston, MA, USA; 2Broad Institute of Harvard and MIT, Cambridge, MA, USA; 3Center for Clinical Epidemiology and Biostatistics and Department of Biostatistics and Epidemiology, University of Pennsylvania, USA; 4 Center for Non-Communicable Diseases, Karachi, Pakistan; 5 Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA; 6Division of Translational Medicine and Human Genetics, Department of Medicine, University of Pennsylvania, USA; 7Department of Public Health and Primary Care, University of Cambridge, UK; 8Department of Genetics, University of Pennsylvania, USA
Learn more about research from Pradeep Natarajan, MD, MMSc
Refer a patient to the Minehan Heart Center